Réaliser un algorithme de détection de faux billets de banque par leurs dimensions :
- Compléter la base de données des données d’apprentissage par une régression linéaire.
- Former l’algorithme d’apprentissage.
- Confronter l’algorithme d’apprentissage et les données des vrais billets.
- Enregistrer le fichier algorithmique.
Notebooks et outils de programmation :




import numpy as np import pandas as pd import statsmodels.api as sm import statsmodels.formula.api as smf import scipy.stats as st from sklearn.linear_model import LinearRegression from sklearn.impute import SimpleImputer import matplotlib.pyplot as plt import seaborn as sns from statsmodels.stats.outliers_influence import variance_inflation_factor import random from sklearn.model_selection import train_test_split import statsmodels.api as sm import pingouin as pg from sklearn import decomposition from sklearn.cluster import KMeans from sklearn.decomposition import PCA import pickle from sklearn.linear_model import LogisticRegression from sklearn.metrics import confusion_matrix from sklearn.naive_bayes import GaussianNB
In [2]:
data = pd.read_csv("billets.csv", sep = ";")
ref = pd.read_csv("billets_production.csv", sep = ",")
In [3]:
data.info() data.describe()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 1500 entries, 0 to 1499 Data columns (total 7 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 is_genuine 1500 non-null bool 1 diagonal 1500 non-null float64 2 height_left 1500 non-null float64 3 height_right 1500 non-null float64 4 margin_low 1463 non-null float64 5 margin_up 1500 non-null float64 6 length 1500 non-null float64 dtypes: bool(1), float64(6) memory usage: 71.9 KB
Out[3]:
| diagonal | height_left | height_right | margin_low | margin_up | length | |
|---|---|---|---|---|---|---|
| count | 1500.000000 | 1500.000000 | 1500.000000 | 1463.000000 | 1500.000000 | 1500.00000 |
| mean | 171.958440 | 104.029533 | 103.920307 | 4.485967 | 3.151473 | 112.67850 |
| std | 0.305195 | 0.299462 | 0.325627 | 0.663813 | 0.231813 | 0.87273 |
| min | 171.040000 | 103.140000 | 102.820000 | 2.980000 | 2.270000 | 109.49000 |
| 25% | 171.750000 | 103.820000 | 103.710000 | 4.015000 | 2.990000 | 112.03000 |
| 50% | 171.960000 | 104.040000 | 103.920000 | 4.310000 | 3.140000 | 112.96000 |
| 75% | 172.170000 | 104.230000 | 104.150000 | 4.870000 | 3.310000 | 113.34000 |
| max | 173.010000 | 104.880000 | 104.950000 | 6.900000 | 3.910000 | 114.44000 |
il y a des valeurs manquantes, identification et compte de ces valeurs :
In [4]:
out_index = data.index[data.isnull().any(axis=1)] print(out_index) print(data.isna().sum())
Int64Index([ 72, 99, 151, 197, 241, 251, 284, 334, 410, 413, 445,
481, 505, 611, 654, 675, 710, 739, 742, 780, 798, 844,
845, 871, 895, 919, 945, 946, 981, 1076, 1121, 1176, 1303,
1315, 1347, 1435, 1438],
dtype='int64')
is_genuine 0
diagonal 0
height_left 0
height_right 0
margin_low 37
margin_up 0
length 0
dtype: int64
Remplacement de ces valeurs par l’utilisation d’une régression linéaire pour prédire les valeurs manquantes.
Répartition des données en 2 datas : l’un avec les données complètes, l’autre avec les données à compléter.
In [5]:
data_in = data.dropna() data_in.info()
<class 'pandas.core.frame.DataFrame'> Int64Index: 1463 entries, 0 to 1499 Data columns (total 7 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 is_genuine 1463 non-null bool 1 diagonal 1463 non-null float64 2 height_left 1463 non-null float64 3 height_right 1463 non-null float64 4 margin_low 1463 non-null float64 5 margin_up 1463 non-null float64 6 length 1463 non-null float64 dtypes: bool(1), float64(6) memory usage: 81.4 KB
In [6]:
data_out = data[data.isnull().any(axis=1)] data_out.info()
<class 'pandas.core.frame.DataFrame'> Int64Index: 37 entries, 72 to 1438 Data columns (total 7 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 is_genuine 37 non-null bool 1 diagonal 37 non-null float64 2 height_left 37 non-null float64 3 height_right 37 non-null float64 4 margin_low 0 non-null float64 5 margin_up 37 non-null float64 6 length 37 non-null float64 dtypes: bool(1), float64(6) memory usage: 2.1 KB
Utilisation d’une régression linéaire pour compléter les données
In [7]:
X = data_in[["height_left", "height_right", "margin_up", "length", "diagonal"]]
In [8]:
y = data_in["margin_low"]
In [9]:
X = sm.add_constant(X) model = sm.OLS(y, X).fit()
In [10]:
model.summary()
Out[10]:
| Dep. Variable: | margin_low | R-squared: | 0.477 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.476 |
| Method: | Least Squares | F-statistic: | 266.1 |
| Date: | Tue, 04 Apr 2023 | Prob (F-statistic): | 2.60e-202 |
| Time: | 09:48:19 | Log-Likelihood: | -1001.3 |
| No. Observations: | 1463 | AIC: | 2015. |
| Df Residuals: | 1457 | BIC: | 2046. |
| Df Model: | 5 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | 22.9948 | 9.656 | 2.382 | 0.017 | 4.055 | 41.935 |
| height_left | 0.1841 | 0.045 | 4.113 | 0.000 | 0.096 | 0.272 |
| height_right | 0.2571 | 0.043 | 5.978 | 0.000 | 0.173 | 0.342 |
| margin_up | 0.2562 | 0.064 | 3.980 | 0.000 | 0.130 | 0.382 |
| length | -0.4091 | 0.018 | -22.627 | 0.000 | -0.445 | -0.374 |
| diagonal | -0.1111 | 0.041 | -2.680 | 0.007 | -0.192 | -0.030 |
| Omnibus: | 73.627 | Durbin-Watson: | 1.893 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 95.862 |
| Skew: | 0.482 | Prob(JB): | 1.53e-21 |
| Kurtosis: | 3.801 | Cond. No. | 1.94e+05 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 1.94e+05. This might indicate that there are
strong multicollinearity or other numerical problems.
La colonne coef sont les coef de la linéarité de la fonction par rapport à chaque donnée P>t est le sinificativité de chacune de ses variables Le résiduel : omnibus, prob omnibus il faudrait que Kurtosis qui doit ête le plus proche de 1
In [11]:
# For each X, calculate VIF and save in dataframe vif = pd.DataFrame() vif["VIF Factor"] = [variance_inflation_factor(X.values, i) for i in range(X.shape[1])] vif["features"] = X.columns
Pas de multicollinéarité dans le modèle
In [12]:
vif
Out[12]:
| VIF Factor | features | |
|---|---|---|
| 0 | 590198.238883 | const |
| 1 | 1.138261 | height_left |
| 2 | 1.230115 | height_right |
| 3 | 1.404404 | margin_up |
| 4 | 1.576950 | length |
| 5 | 1.013613 | diagonal |
In [13]:
ax = pg.qqplot(model.resid, dist = "norm")
#plt.savefig("01.jpg", dpi = 150)
/Users/photos/opt/anaconda3/lib/python3.9/site-packages/outdated/utils.py:14: OutdatedPackageWarning: The package pingouin is out of date. Your version is 0.5.2, the latest is 0.5.3. Set the environment variable OUTDATED_IGNORE=1 to disable these warnings. return warn( /Users/photos/opt/anaconda3/lib/python3.9/site-packages/outdated/utils.py:14: OutdatedPackageWarning: The package outdated is out of date. Your version is 0.2.1, the latest is 0.2.2. Set the environment variable OUTDATED_IGNORE=1 to disable these warnings. return warn(
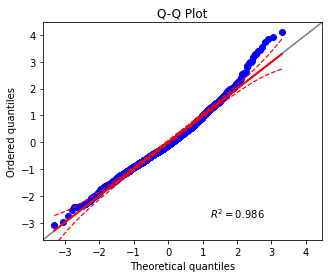
Création d’un modèle de régression linéaire
In [14]:
regression_model = LinearRegression()
Ajustement du modèle aux données connues
In [15]:
X
Out[15]:
| const | height_left | height_right | margin_up | length | diagonal | |
|---|---|---|---|---|---|---|
| 0 | 1.0 | 104.86 | 104.95 | 2.89 | 112.83 | 171.81 |
| 1 | 1.0 | 103.36 | 103.66 | 2.99 | 113.09 | 171.46 |
| 2 | 1.0 | 104.48 | 103.50 | 2.94 | 113.16 | 172.69 |
| 3 | 1.0 | 103.91 | 103.94 | 3.01 | 113.51 | 171.36 |
| 4 | 1.0 | 104.28 | 103.46 | 3.48 | 112.54 | 171.73 |
| … | … | … | … | … | … | … |
| 1495 | 1.0 | 104.38 | 104.17 | 3.09 | 111.28 | 171.75 |
| 1496 | 1.0 | 104.63 | 104.44 | 3.37 | 110.97 | 172.19 |
| 1497 | 1.0 | 104.01 | 104.12 | 3.36 | 111.95 | 171.80 |
| 1498 | 1.0 | 104.28 | 104.06 | 3.46 | 112.25 | 172.06 |
| 1499 | 1.0 | 104.15 | 103.82 | 3.37 | 112.07 | 171.47 |
1463 rows × 6 columns
In [16]:
regression_model.fit(X[X.columns[1:]], y)
Out[16]:
LinearRegression()
Sélection des observations avec des valeurs manquantes
In [17]:
missing_data = data[data["margin_low"].isnull()]
Sélection des variables à utiliser pour prédire les valeurs manquantes
In [18]:
X_missing = missing_data[["height_left", "height_right", "margin_up", "length", "diagonal"]]
Prédiction des valeurs manquantes
In [19]:
y_missing = regression_model.predict(X_missing)
Remplacement des valeurs manquantes dans la base de données
In [20]:
data.loc[data["margin_low"].isnull(), "margin_low"] = y_missing data.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 1500 entries, 0 to 1499 Data columns (total 7 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 is_genuine 1500 non-null bool 1 diagonal 1500 non-null float64 2 height_left 1500 non-null float64 3 height_right 1500 non-null float64 4 margin_low 1500 non-null float64 5 margin_up 1500 non-null float64 6 length 1500 non-null float64 dtypes: bool(1), float64(6) memory usage: 71.9 KB
Les données sont complétées
In [21]:
compar_1 = []
for i in out_index:
compar_1.append(data["margin_low"].iloc[i])
compar_1.sort()
In [22]:
#not_index = [] #while len(not_index) < 37: # num = random.randint(1, 1500) # if num not in out_index and num not in not_index: # not_index.append(num)
In [23]:
#np.array(not_index)
In [24]:
not_index = [ 473, 285, 1234, 1229, 214, 392, 1481, 416, 440, 1363, 220,
1085, 247, 553, 571, 1035, 437, 1370, 974, 1322, 792, 907,
1231, 1100, 232, 706, 1003, 790, 803, 1369, 433, 1388, 537,
1349, 1237, 658, 346]
In [25]:
compar_2 = []
for i in not_index:
compar_2.append(data["margin_low"].iloc[i])
compar_2.sort()
Comparaison en données existantes et données remplacées
In [26]:
comparaison = pd.DataFrame(compar_2, compar_1).reset_index().rename(columns = {"index" : "Remplacées", 0 : "Existantes"})
comparaison
Out[26]:
| Remplacées | Existantes | |
|---|---|---|
| 0 | 3.614306 | 3.25 |
| 1 | 3.746333 | 3.75 |
| 2 | 3.768554 | 3.77 |
| 3 | 3.803308 | 3.85 |
| 4 | 3.893748 | 3.85 |
| 5 | 4.058764 | 3.90 |
| 6 | 4.080414 | 3.93 |
| 7 | 4.093621 | 3.94 |
| 8 | 4.094065 | 4.04 |
| 9 | 4.127442 | 4.10 |
| 10 | 4.135034 | 4.11 |
| 11 | 4.137780 | 4.15 |
| 12 | 4.160539 | 4.25 |
| 13 | 4.160607 | 4.29 |
| 14 | 4.177420 | 4.31 |
| 15 | 4.179736 | 4.38 |
| 16 | 4.237415 | 4.43 |
| 17 | 4.249629 | 4.47 |
| 18 | 4.298047 | 4.51 |
| 19 | 4.318525 | 4.51 |
| 20 | 4.319014 | 4.53 |
| 21 | 4.341643 | 4.54 |
| 22 | 4.371811 | 4.60 |
| 23 | 4.393668 | 4.72 |
| 24 | 4.410457 | 4.84 |
| 25 | 4.439846 | 4.86 |
| 26 | 4.470650 | 5.06 |
| 27 | 4.650617 | 5.19 |
| 28 | 4.710533 | 5.27 |
| 29 | 4.778967 | 5.38 |
| 30 | 4.802145 | 5.40 |
| 31 | 5.047570 | 5.46 |
| 32 | 5.050277 | 5.52 |
| 33 | 5.067584 | 5.77 |
| 34 | 5.140043 | 5.80 |
| 35 | 5.185862 | 6.05 |
| 36 | 5.726993 | 6.19 |
Remplacement validé. La plus petite valeur remplacée est plus grande la plus petite valeur existante et la plus grande valeur remplacée est plus petite que la plus grande valeur existante.
Description des variables
In [27]:
desc = data.describe()
Analyse univariée des différentes variables avec des box plot
In [28]:
for col in data.columns[1:]:
plt.figure()
plt.boxplot(x = data[col])
plt.title(col)
# plt.savefig(f"02 {col} .jpg", dpi = 150)
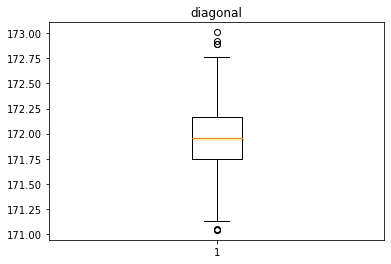
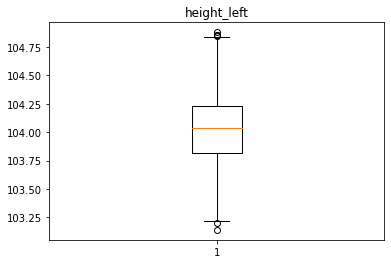
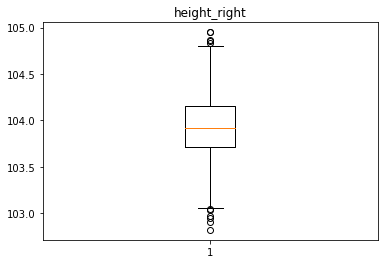
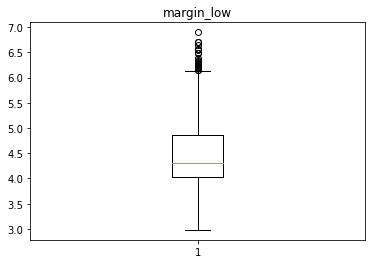
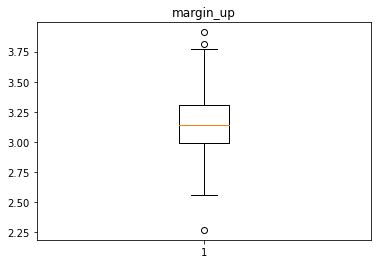

Identification des valeurs aberrantes
In [29]:
outliers = pd.DataFrame(columns = data.columns)
for col in data.columns:
q1 = desc.loc['25%', ["diagonal", "height_left", "height_right", "margin_low", "margin_up", "length"]]
q3 = desc.loc['75%', ["diagonal", "height_left", "height_right", "margin_low", "margin_up", "length"]]
iqr = q3 - q1
lower_bound = q1 - 1.5 * iqr
upper_bound = q3 + 1.5 * iqr
outliers[["diagonal", "height_left", "height_right", "margin_low", "margin_up", "length"]] = data[(data[1:] < lower_bound) | (data[1:] > upper_bound)][["diagonal", "height_left", "height_right", "margin_low", "margin_up", "length"]]
/var/folders/0p/7nmt36t11y5fcm_wdkxslb000000gn/T/ipykernel_1013/754005521.py:8: FutureWarning: Automatic reindexing on DataFrame vs Series comparisons is deprecated and will raise ValueError in a future version. Do `left, right = left.align(right, axis=1, copy=False)` before e.g. `left == right` outliers[["diagonal", "height_left", "height_right", "margin_low", "margin_up", "length"]] = data[(data[1:] < lower_bound) | (data[1:] > upper_bound)][["diagonal", "height_left", "height_right", "margin_low", "margin_up", "length"]]
Étude de la distribution
In [30]:
for col in data.columns[1:]:
plt.figure()
plt.hist(data[col], bins = 20)
plt.title(col)
# plt.savefig(f"03 {col} .jpg", dpi = 150)
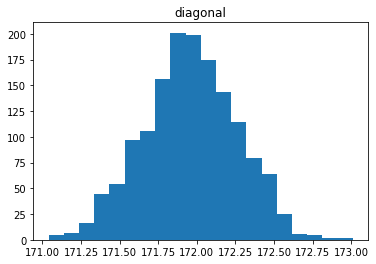
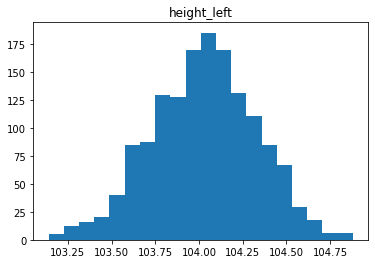

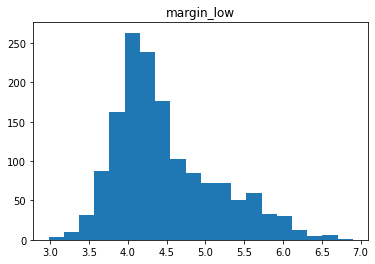
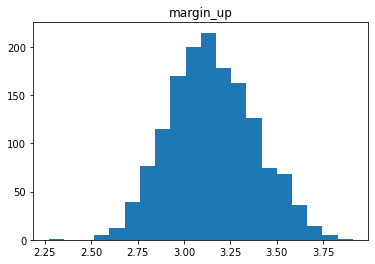

Analyse des relations entre les variables
In [31]:
corr = data.corr()
plt.figure()
plt.imshow(corr, cmap = 'coolwarm')
plt.colorbar()
plt.xticks(range(len(corr.columns[1:])), corr.columns[1:], rotation = 90)
plt.yticks(range(len(corr.columns[1:])), corr.columns[1:])
plt.title("Relations entre variables", fontsize = 15)
#plt.savefig("04.jpg", dpi = 150)
plt.show()

Pairplot en fonction du biais
In [32]:
figg = sns.pairplot(data, hue = "is_genuine")
figg.fig.suptitle("Affichage par variable en fonction du biais", fontsize = 20, y = 1.02);
#plt.savefig("05.jpg", dpi = 150)
Il faut séparer les données en 2 parties avec une équivalente répartition entre valeurs remplacées et valeurs d’origine
Séparation en 2 fichiers
In [33]:
data.head()
Out[33]:
| is_genuine | diagonal | height_left | height_right | margin_low | margin_up | length | |
|---|---|---|---|---|---|---|---|
| 0 | True | 171.81 | 104.86 | 104.95 | 4.52 | 2.89 | 112.83 |
| 1 | True | 171.46 | 103.36 | 103.66 | 3.77 | 2.99 | 113.09 |
| 2 | True | 172.69 | 104.48 | 103.50 | 4.40 | 2.94 | 113.16 |
| 3 | True | 171.36 | 103.91 | 103.94 | 3.62 | 3.01 | 113.51 |
| 4 | True | 171.73 | 104.28 | 103.46 | 4.04 | 3.48 | 112.54 |
In [34]:
X_train, X_test, y_train, y_test = train_test_split(data.drop(columns = "is_genuine", axis = 1), data["is_genuine"], test_size = 0.33, stratify = data["is_genuine"], random_state = 42)
Sélection de faux billets pour l’apprentissage
In [35]:
X_train.head()
Out[35]:
| diagonal | height_left | height_right | margin_low | margin_up | length | |
|---|---|---|---|---|---|---|
| 780 | 172.41 | 103.95 | 103.79 | 4.080414 | 3.13 | 113.41 |
| 1367 | 171.60 | 104.37 | 104.20 | 5.820000 | 3.08 | 112.84 |
| 1477 | 172.16 | 104.23 | 104.19 | 5.090000 | 3.61 | 112.43 |
| 1203 | 172.02 | 104.22 | 104.19 | 5.140000 | 3.73 | 110.49 |
| 1192 | 171.95 | 104.08 | 104.08 | 5.660000 | 3.32 | 110.93 |
In [36]:
X_test.head()
Out[36]:
| diagonal | height_left | height_right | margin_low | margin_up | length | |
|---|---|---|---|---|---|---|
| 41 | 172.08 | 104.19 | 103.82 | 3.99 | 3.21 | 113.20 |
| 426 | 171.91 | 103.99 | 103.50 | 3.41 | 2.92 | 113.02 |
| 132 | 171.84 | 103.77 | 103.98 | 4.61 | 2.99 | 113.59 |
| 1295 | 171.90 | 103.97 | 104.36 | 5.59 | 3.61 | 112.05 |
| 360 | 171.61 | 103.73 | 104.01 | 4.29 | 2.93 | 112.76 |
In [37]:
#X_test.to_csv("file_test.csv", index = False)
Comptage des True et False
In [38]:
y_train = data.loc[:, data.columns == "is_genuine"] y_train.value_counts()
Out[38]:
is_genuine True 1000 False 500 dtype: int64
In [39]:
X_train = data.loc[:, data.columns != "is_genuine"] X_train = sm.tools.add_constant(X_train) X_train.head()
Out[39]:
| const | diagonal | height_left | height_right | margin_low | margin_up | length | |
|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 171.81 | 104.86 | 104.95 | 4.52 | 2.89 | 112.83 |
| 1 | 1.0 | 171.46 | 103.36 | 103.66 | 3.77 | 2.99 | 113.09 |
| 2 | 1.0 | 172.69 | 104.48 | 103.50 | 4.40 | 2.94 | 113.16 |
| 3 | 1.0 | 171.36 | 103.91 | 103.94 | 3.62 | 3.01 | 113.51 |
| 4 | 1.0 | 171.73 | 104.28 | 103.46 | 4.04 | 3.48 | 112.54 |
Prédiction avec k-means en utilisant 2 clusters
In [40]:
# Nombre de clusters: n_clust = 2 # Clustering par K-means: km = KMeans(n_clusters = n_clust,random_state = 1994) x_km = km.fit_transform(data[["diagonal","height_left","height_right","margin_low","margin_up","length"]])
In [41]:
# Ajout d'une colonne contenant le cluster clusters_km = km.labels_
In [42]:
data["cluster_km"] = km.labels_ data["cluster_km"] = data["cluster_km"].apply(str)
In [43]:
centroids_km = km.cluster_centers_
In [44]:
# Clustering par projection des individus sur le premier plan factoriel: pca_km = decomposition.PCA(n_components = 3).fit(data[["diagonal","height_left","height_right","margin_low","margin_up","length"]]) acp_km = PCA(n_components = 3).fit_transform(data[["diagonal","height_left","height_right","margin_low","margin_up","length"]]) centroids_km_projected = pca_km.transform(centroids_km)
In [45]:
# Graphique:
for couleur,k in zip(["#AAAAAA", "#55AA55"],[0,1]):
plt.scatter(acp_km[km.labels_ == k, 0], acp_km[km.labels_ == k, 1], c = couleur, s = 60, edgecolors="#FFFFFF", label = "Cluster {}".format(k))
plt.legend()
plt.scatter(centroids_km_projected[:, 0],centroids_km_projected[:, 1], color="blue", label="Centroïdes")
plt.title("Projection des individus et des {} centroïdes sur le premier plan factoriel".format(len(centroids_km)), fontsize = 10)
#plt.savefig("06.jpg", dpi = 150)
plt.show()
#Vérification de la classification: Matrice de confusion:
km_matrix = pd.crosstab(data["is_genuine"], clusters_km)
print(km_matrix)
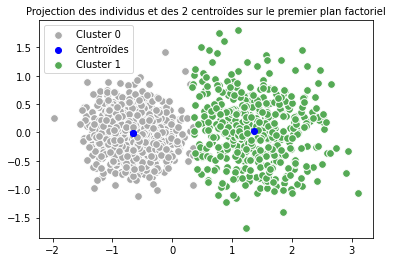
col_0 0 1 is_genuine False 19 481 True 997 3
In [46]:
plt.title("Projection des point en fonction de leur véritable nature")
sns.scatterplot(x = acp_km[:, 0], y = acp_km[:, 1], hue = data["is_genuine"]);
#plt.savefig("06b.jpg", dpi = 150)
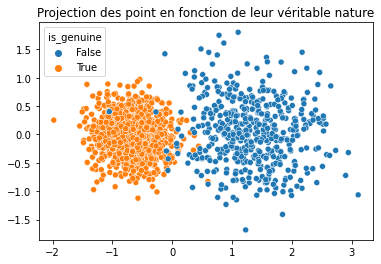
In [47]:
# Graphique:
plt.figure(figsize=(16, 6))
plt.subplot(1, 2, 1)
sns.heatmap(km_matrix,
annot = True,
fmt = ".3g",
cmap = sns.color_palette("mako", as_cmap = True),
linecolor = "white",
linewidths = 0.3,
xticklabels = ["1", "0"],
yticklabels = ["Faux","Vrai"])
plt.xlabel("Cluster")
plt.ylabel("is_genuin")
plt.title("Matrice de confusion", fontsize = 15)
plt.subplot(1, 2, 2)
for couleur,k in zip(["#AAAAAA", "#55AA55"],[0,1]):
plt.scatter(acp_km[km.labels_ == k, 0],acp_km[km.labels_ == k, 1],c = couleur, s = 60, edgecolors="#FFFFFF", label = "Cluster {}".format(k))
plt.legend()
plt.scatter(centroids_km_projected[:, 0],centroids_km_projected[:, 1], color = "blue", label="Centroïdes")
plt.title("Projection des individus et des {} centroïdes sur le premier plan factoriel".format(len(centroids_km)), fontsize = 15)
#plt.savefig("07.jpg", dpi = 150)
;
Out[47]:
''

In [48]:
data.head()
Out[48]:
| is_genuine | diagonal | height_left | height_right | margin_low | margin_up | length | cluster_km | |
|---|---|---|---|---|---|---|---|---|
| 0 | True | 171.81 | 104.86 | 104.95 | 4.52 | 2.89 | 112.83 | 0 |
| 1 | True | 171.46 | 103.36 | 103.66 | 3.77 | 2.99 | 113.09 | 0 |
| 2 | True | 172.69 | 104.48 | 103.50 | 4.40 | 2.94 | 113.16 | 0 |
| 3 | True | 171.36 | 103.91 | 103.94 | 3.62 | 3.01 | 113.51 | 0 |
| 4 | True | 171.73 | 104.28 | 103.46 | 4.04 | 3.48 | 112.54 | 0 |
Régression logistique
In [49]:
# Construction du modèle et ajustement des données log_reg = sm.Logit(y_train, X_train).fit()
Optimization terminated successfully.
Current function value: 0.028228
Iterations 13
In [50]:
log_reg.summary()
Out[50]:
| Dep. Variable: | is_genuine | No. Observations: | 1500 |
|---|---|---|---|
| Model: | Logit | Df Residuals: | 1493 |
| Method: | MLE | Df Model: | 6 |
| Date: | Tue, 04 Apr 2023 | Pseudo R-squ.: | 0.9557 |
| Time: | 09:48:33 | Log-Likelihood: | -42.342 |
| converged: | True | LL-Null: | -954.77 |
| Covariance Type: | nonrobust | LLR p-value: | 0.000 |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| const | -204.5582 | 241.768 | -0.846 | 0.398 | -678.415 | 269.299 |
| diagonal | 0.0680 | 1.091 | 0.062 | 0.950 | -2.071 | 2.207 |
| height_left | -1.7162 | 1.104 | -1.555 | 0.120 | -3.880 | 0.447 |
| height_right | -2.2584 | 1.072 | -2.107 | 0.035 | -4.359 | -0.157 |
| margin_low | -5.7756 | 0.937 | -6.164 | 0.000 | -7.612 | -3.939 |
| margin_up | -10.1531 | 2.108 | -4.817 | 0.000 | -14.284 | -6.022 |
| length | 5.9129 | 0.846 | 6.991 | 0.000 | 4.255 | 7.571 |
Possibly complete quasi-separation: A fraction 0.51 of observations can be
perfectly predicted. This might indicate that there is complete
quasi-separation. In this case some parameters will not be identified.
In [51]:
clf = LogisticRegression(random_state=0).fit(X_train[["height_right", "margin_low", "margin_up", "length"]], y_train)
/Users/photos/opt/anaconda3/lib/python3.9/site-packages/sklearn/utils/validation.py:63: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel(). return f(*args, **kwargs)
In [52]:
prediction = clf.predict(X_test[["height_right", "margin_low", "margin_up", "length"]]) prediction
Out[52]:
array([ True, True, True, False, True, True, True, True, True,
False, True, True, True, False, True, True, True, False,
False, False, False, False, True, False, False, True, True,
False, True, True, False, True, True, False, False, True,
True, True, True, True, True, True, True, True, False,
True, False, True, False, True, False, False, False, True,
True, True, True, True, True, True, False, True, True,
True, True, False, False, True, True, False, True, True,
True, True, True, True, True, False, False, True, True,
True, True, False, True, False, True, True, False, True,
True, True, True, True, False, True, False, False, False,
True, False, True, False, True, True, False, True, True,
False, True, True, False, False, False, True, True, False,
True, True, True, True, True, True, True, False, False,
True, True, False, True, False, False, False, True, True,
True, False, True, True, True, True, True, True, True,
True, True, True, True, False, False, True, True, False,
False, True, True, False, False, True, True, True, False,
True, True, True, False, True, True, True, False, True,
True, True, False, True, True, True, True, True, False,
True, True, True, False, False, False, True, True, False,
True, False, True, False, True, True, True, False, True,
True, True, True, False, False, True, False, True, False,
True, True, True, True, False, False, True, True, True,
True, False, False, True, False, False, True, True, True,
False, True, True, True, True, True, True, True, False,
True, True, True, True, False, True, False, True, True,
True, True, False, True, True, True, True, False, False,
True, True, False, True, True, True, True, False, True,
True, True, False, True, True, False, False, False, False,
True, False, False, False, True, True, True, True, False,
True, True, True, True, False, True, True, True, False,
False, True, True, True, True, False, True, False, True,
False, False, False, False, True, False, True, True, True,
True, False, False, False, False, True, True, True, True,
True, True, False, True, False, False, True, True, True,
True, False, True, True, True, False, True, True, False,
True, True, False, True, True, False, False, True, True,
True, True, True, True, False, False, True, True, False,
True, True, True, True, False, True, True, True, True,
True, True, True, True, True, False, True, False, True,
True, True, True, False, True, True, True, False, True,
True, False, True, True, True, False, True, True, True,
True, False, False, False, True, True, True, True, True,
True, False, False, True, True, True, False, True, True,
True, True, True, False, False, True, False, True, False,
True, False, True, True, True, False, False, False, True,
False, True, True, True, True, False, True, True, True,
True, True, True, True, True, True, True, True, False,
False, False, False, False, False, False, True, True, False,
True, True, False, False, True, True, False, False, True,
True, True, True, True, True, True, False, True, False,
False, False, True, True, False, True, True, True, True,
True, True, True, True, False, False, True, True, True,
True, True, True, True, True, False, False, True, True])
In [53]:
X_train
Out[53]:
| const | diagonal | height_left | height_right | margin_low | margin_up | length | |
|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 171.81 | 104.86 | 104.95 | 4.52 | 2.89 | 112.83 |
| 1 | 1.0 | 171.46 | 103.36 | 103.66 | 3.77 | 2.99 | 113.09 |
| 2 | 1.0 | 172.69 | 104.48 | 103.50 | 4.40 | 2.94 | 113.16 |
| 3 | 1.0 | 171.36 | 103.91 | 103.94 | 3.62 | 3.01 | 113.51 |
| 4 | 1.0 | 171.73 | 104.28 | 103.46 | 4.04 | 3.48 | 112.54 |
| … | … | … | … | … | … | … | … |
| 1495 | 1.0 | 171.75 | 104.38 | 104.17 | 4.42 | 3.09 | 111.28 |
| 1496 | 1.0 | 172.19 | 104.63 | 104.44 | 5.27 | 3.37 | 110.97 |
| 1497 | 1.0 | 171.80 | 104.01 | 104.12 | 5.51 | 3.36 | 111.95 |
| 1498 | 1.0 | 172.06 | 104.28 | 104.06 | 5.17 | 3.46 | 112.25 |
| 1499 | 1.0 | 171.47 | 104.15 | 103.82 | 4.63 | 3.37 | 112.07 |
1500 rows × 7 columns
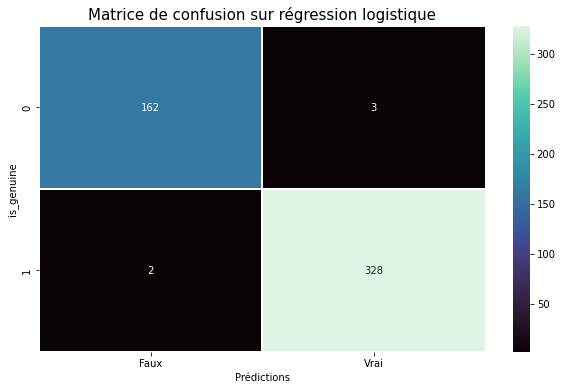
Matrice de confusion
In [54]:
# Créer la matrice de confusion confusion_matrix = confusion_matrix(y_test, prediction) print(confusion_matrix)
[[162 3] [ 2 328]]
In [55]:
# Graphique:
plt.figure(figsize=(10, 6))
plt.subplot()
sns.heatmap(confusion_matrix,
annot = True,
fmt = ".3g",
cmap = sns.color_palette("mako", as_cmap = True),
linecolor = "white",
linewidths = 0.3,
xticklabels = ["Faux","Vrai"],
yticklabels = ["0", "1"])
plt.xlabel("Prédictions")
plt.ylabel("is_genuine")
plt.title("Matrice de confusion sur régression logistique", fontsize = 15);
#plt.savefig("08.jpg", dpi = 150)

In [56]:
ref
Out[56]:
| diagonal | height_left | height_right | margin_low | margin_up | length | id | |
|---|---|---|---|---|---|---|---|
| 0 | 171.76 | 104.01 | 103.54 | 5.21 | 3.30 | 111.42 | A_1 |
| 1 | 171.87 | 104.17 | 104.13 | 6.00 | 3.31 | 112.09 | A_2 |
| 2 | 172.00 | 104.58 | 104.29 | 4.99 | 3.39 | 111.57 | A_3 |
| 3 | 172.49 | 104.55 | 104.34 | 4.44 | 3.03 | 113.20 | A_4 |
| 4 | 171.65 | 103.63 | 103.56 | 3.77 | 3.16 | 113.33 | A_5 |
In [57]:
# Predictions sur des donnees inconnues: X_test_clf = ref[["height_right","margin_low","margin_up","length"]] ref["reg_pred"] = clf.predict(X_test_clf) print(ref[["id","reg_pred"]])
id reg_pred 0 A_1 False 1 A_2 False 2 A_3 False 3 A_4 True 4 A_5 True
In [58]:
ref
Out[58]:
| diagonal | height_left | height_right | margin_low | margin_up | length | id | reg_pred | |
|---|---|---|---|---|---|---|---|---|
| 0 | 171.76 | 104.01 | 103.54 | 5.21 | 3.30 | 111.42 | A_1 | False |
| 1 | 171.87 | 104.17 | 104.13 | 6.00 | 3.31 | 112.09 | A_2 | False |
| 2 | 172.00 | 104.58 | 104.29 | 4.99 | 3.39 | 111.57 | A_3 | False |
| 3 | 172.49 | 104.55 | 104.34 | 4.44 | 3.03 | 113.20 | A_4 | True |
| 4 | 171.65 | 103.63 | 103.56 | 3.77 | 3.16 | 113.33 | A_5 | True |
In [59]:
#file_export = ref[["diagonal", "height_left", "height_right", "margin_low", "margin_up", "length", "id"]]
In [60]:
#file_export.to_csv("ref_predict.csv", index = False)
In [61]:
# save
#with open("model.pkl", "wb") as f:
# pickle.dump(clf,f)
In [ ]:
Direction vers l’application de détection de billets.
Retour vers Data-Analyst
